3: Immune Infiltration Overlap Analysis
Demonstrate the usage of SpatialCells to analyse Immune cell infiltration into the tumor region
@author: Guihong Wan and Boshen Yan
@date: Sept 8, 2023
@last updated: Oct 6, 2023
[6]:
import pandas as pd
import matplotlib.pyplot as plt
import anndata as ad
import spatialcells as spc
Read and preprocess data
[13]:
adata = ad.read_h5ad("../../data/MEL1_adata.h5ad")
spc.prep.setGate(adata, "KERATIN_cellRingMask", 6.4, debug=True)
spc.prep.setGate(adata, "SOX10_cellRingMask", 7.9, debug=True)
spc.prep.setGate(adata, "CD3D_cellRingMask", 7, debug=True)
KERATIN_cellRingMask_positive
False 1067400
True 43185
Name: count, dtype: int64
SOX10_cellRingMask_positive
False 566576
True 544009
Name: count, dtype: int64
CD3D_cellRingMask_positive
False 1038559
True 72026
Name: count, dtype: int64
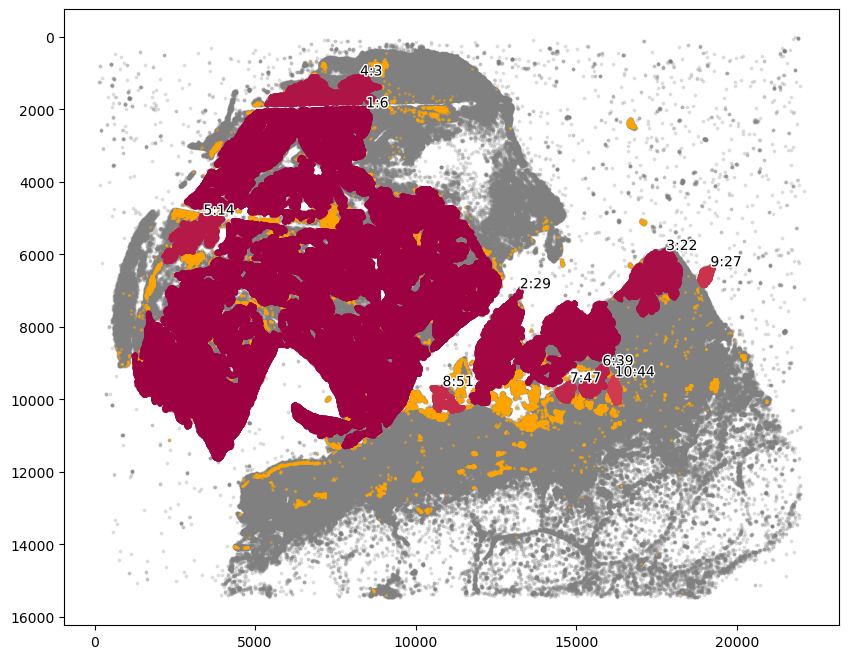
Isolate tumor cell communities and draw region boundary as before
[14]:
marker = ["SOX10_cellRingMask_positive"]
communitycolumn = "COI_community"
ret = spc.spatial.getCommunities(adata, marker, eps=60, newcolumn=communitycolumn)
fig, ax = plt.subplots(figsize=(10, 8))
spc.plt.plotCommunities(
adata, ret, communitycolumn, plot_first_n_clusters=10, s=2, fontsize=10, ax=ax
)
ax.invert_yaxis()
plt.show()
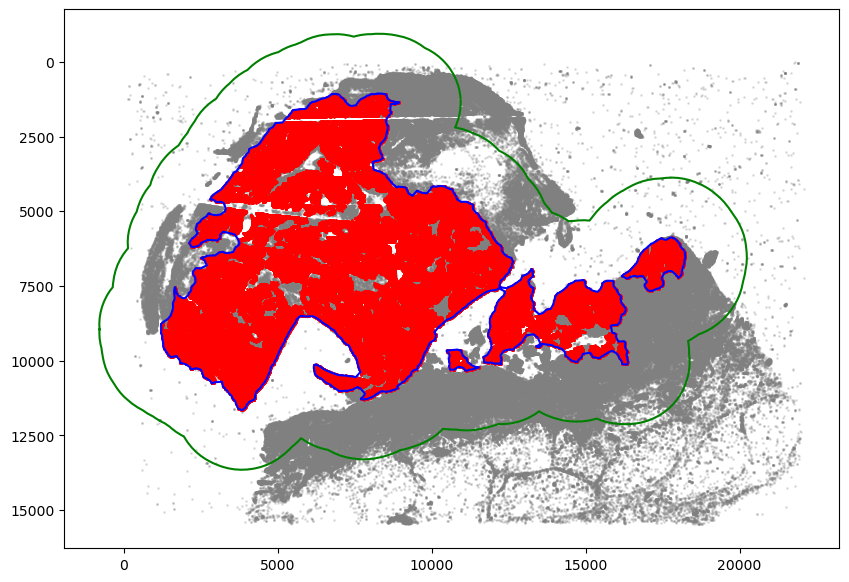
[15]:
communityIndexList = [6, 3, 14, 51, 29, 47, 39, 44, 22]
boundary = spc.spatial.getBoundary(
adata, communitycolumn, communityIndexList, alpha=130
)
boundary = spc.spa.pruneSmallComponents(boundary, min_edges=50, holes_min_edges=500)
roi_boundary = spc.spa.getExtendedBoundary(boundary, offset=2000)
markersize = 1
fig, ax = plt.subplots(figsize=(10, 7))
## all points
ax.scatter(
*zip(*adata.obs[["X_centroid", "Y_centroid"]].to_numpy()),
s=markersize,
color="grey",
alpha=0.2
)
# Points in selected commnities
xy = adata.obs[adata.obs[communitycolumn].isin(communityIndexList)][
["X_centroid", "Y_centroid"]
].to_numpy()
ax.scatter(xy[:, 0], xy[:, 1], s=markersize, color="r")
# Bounds of points in selected commnities
spc.plt.plotBoundary(boundary, ax=ax, label="Boundary", color="b")
spc.plt.plotBoundary(roi_boundary, ax=ax, label="ROI boundary", color="g")
ax.invert_yaxis()
plt.show()
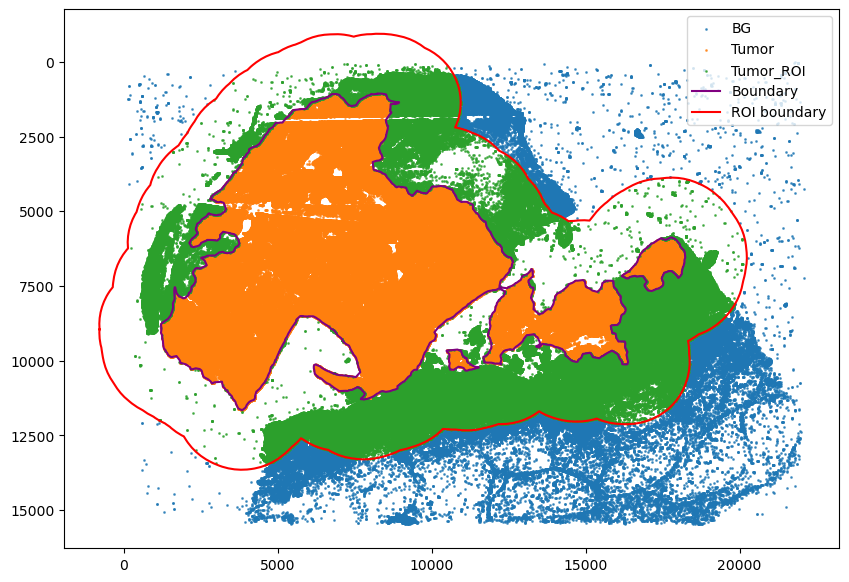
Assign cells to tumor region
[18]:
spc.spatial.assignPointsToRegions(
adata,
[boundary, roi_boundary],
["Tumor", "Tumor_ROI"],
assigncolumn="region",
default="BG",
)
point_size = 1
fig, ax = plt.subplots(figsize=(10, 7))
for region in sorted(set(adata.obs["region"])):
tmp = adata.obs[adata.obs.region == region]
ax.scatter(
*zip(*tmp[["X_centroid", "Y_centroid"]].to_numpy()),
s=point_size,
alpha=0.7,
label=region
)
# Bounds of points in selected commnities
spc.plt.plotBoundary(boundary, ax=ax, label="Boundary", color="purple")
spc.plt.plotBoundary(roi_boundary, ax=ax, label="ROI boundary", color="r")
plt.legend(loc="upper right")
ax.invert_yaxis()
plt.show()
955899it [02:32, 6267.29it/s]
Assigned points to region: Tumor
403552it [00:18, 22222.54it/s]
Assigned points to region: Tumor_ROI
Generalize cell-types based on existing phenotypes
[19]:
def merge_pheno(row):
if row["phenotype_large_cohort"] in [
"T cells",
"Cytotoxic T cells",
"Exhausted T cells",
]:
return "T cells"
elif row["phenotype_large_cohort"] in ["Melanocytes"]:
return "Tumor cells"
else:
return "Other cells"
def cell_type(row):
if row["SOX10_cellRingMask_positive"]:
return "SOX10+"
elif row["CD3D_cellRingMask_positive"]:
return "CD3D+"
else:
return "Other cells"
# Applying the function to create the new columns
adata.obs["pheno1"] = pd.Categorical(adata.obs.apply(merge_pheno, axis=1))
adata.obs["Cell Types"] = pd.Categorical(adata.obs.apply(cell_type, axis=1))
[20]:
spc.msmt.getRegionComposition(adata, "pheno1")
[20]:
| pheno1 | cell_count | composition | |
|---|---|---|---|
| 0 | Tumor cells | 524293 | 0.472087 |
| 1 | Other cells | 516082 | 0.464694 |
| 2 | T cells | 70210 | 0.063219 |
Find immune cell infiltrated areas in the tumor ROI region
[23]:
melano = adata[
(adata.obs.SOX10_cellRingMask_positive) & (adata.obs.region.isin(["Tumor_ROI", "Tumor"]))
]
tcells = adata[
(adata.obs.CD3D_cellRingMask_positive)
& (adata.obs.region.isin(["Tumor_ROI", "Tumor"]))
]
fig, ax = plt.subplots(figsize=(10, 7))
ax.invert_yaxis()
ax.set_aspect("equal")
plt.scatter(
tcells.obs["X_centroid"],
tcells.obs["Y_centroid"],
s=0.5,
label="T cells",
color="green",
alpha=0.5,
)
spc.plt.plotBoundary(roi_boundary, ax=ax, label="ROI boundary", color="r")
plt.legend(loc="upper right", markerscale=5)
plt.show()
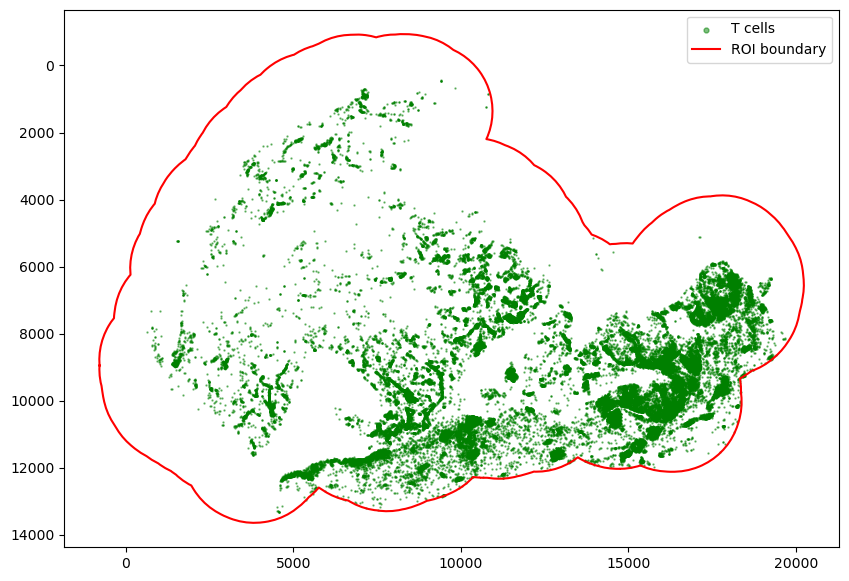
[25]:
tumor = adata[adata.obs.region.isin(["Tumor_ROI", "Tumor"])]
communitycolumn = "CD3D_cellRingMask_positive"
communityIndexList = [True]
immune_boundary = spc.spatial.getBoundary(
tumor, communitycolumn, communityIndexList, alpha=130
)
immune_boundary = spc.spa.pruneSmallComponents(
immune_boundary, min_edges=25, holes_min_edges=30, min_area=30000
)
[26]:
markersize = 0.1
fig, ax = plt.subplots(figsize=(10, 7))
## all points
ax.scatter(
*zip(*adata.obs[["X_centroid", "Y_centroid"]].to_numpy()),
s=markersize,
color="grey",
alpha=0.2
)
# Points in selected commnities
xy = tumor.obs[tumor.obs[communitycolumn].isin(communityIndexList)][
["X_centroid", "Y_centroid"]
].to_numpy()
ax.scatter(xy[:, 0], xy[:, 1], s=markersize, color="green", alpha=1, label="T cells")
# Bounds of points in selected commnities
spc.plt.plotBoundary(
immune_boundary, ax=ax, label="Immune Cell Region Boundary", color="k", linewidth=1
)
spc.plt.plotBoundary(roi_boundary, ax=ax, label="ROI boundary", color="r")
# ax.set_xlim(0, 20000)
# ax.set_ylim(0, 13000)
ax.invert_yaxis()
ax.set_aspect("equal")
ax.set_axis_off()
# plt.legend(loc="upper right", markerscale=5, fontsize=13.5)
# plt.savefig("immune_cell_region1.png", dpi=400)
plt.show()
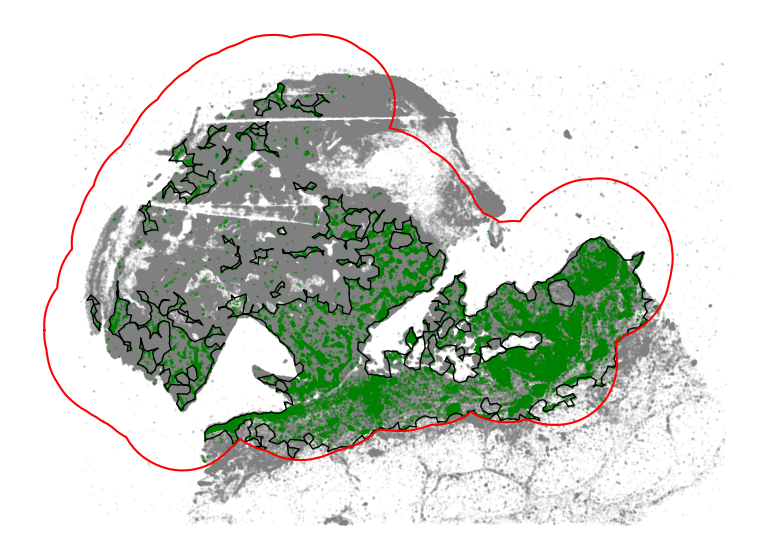
[27]:
spc.spatial.assignPointsToRegions(
melano, [immune_boundary], ["T"], assigncolumn="tumor_isolated_region", default="F"
)
543014it [06:55, 1305.99it/s]
Assigned points to region: T
We can estimate the scale of immune infiltration by overlapping the identified immune infiltrated regions with all tumor cells.
[28]:
point_size = 0.5
fig, ax = plt.subplots(figsize=(10, 7))
ax.scatter(
*zip(*adata.obs[["X_centroid", "Y_centroid"]].to_numpy()),
s=markersize,
color="grey",
alpha=0.2
)
colors = ["red", "orange"]
labels = ["Immune-isolated Tumor Cells", "Immune-rich Tumor Cells"]
for i, region in enumerate(sorted(set(melano.obs["tumor_isolated_region"]))):
tmp = melano.obs[melano.obs.tumor_isolated_region == region]
ax.scatter(
*zip(*tmp[["X_centroid", "Y_centroid"]].to_numpy()),
s=point_size,
alpha=0.5,
color=colors[i],
label=labels[i]
)
# Bounds of points in selected commnities
spc.plt.plotBoundary(
immune_boundary, ax=ax, label="Immune Cell Region Boundary", color="k", linewidth=1
)
spc.plt.plotBoundary(roi_boundary, ax=ax, label="ROI boundary", color="b")
ax.invert_yaxis()
ax.set_axis_off()
# plt.savefig("roi_region1.png", dpi=400)
plt.show()
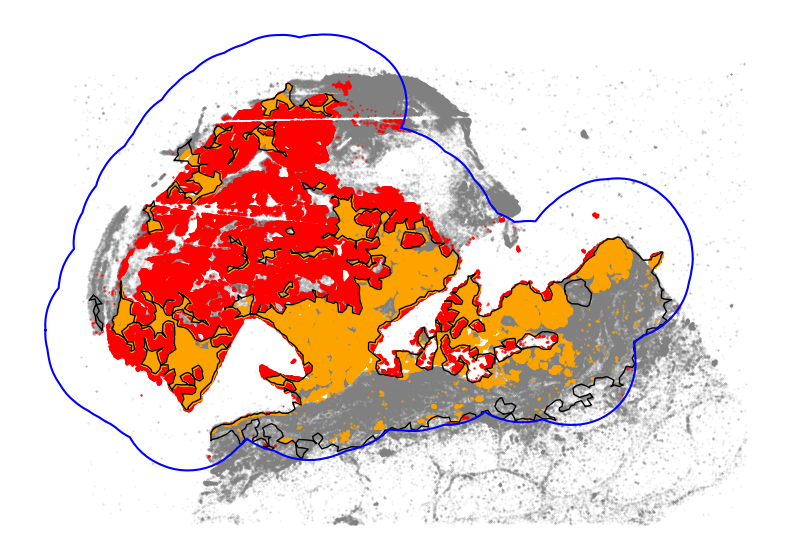
[30]:
print("Percentage of tumor cells in immune-isolated regions: ")
melano.obs["tumor_isolated_region"].value_counts() / len(melano.obs)
Percentage of tumor cells in immune-isolated regions:
[30]:
tumor_isolated_region
F 0.580213
T 0.419787
Name: count, dtype: float64
We can also look at the scale of immune infiltration by comparing the area overlap between tumor regions and immune cell regions.
[31]:
roi_area = spc.msmt.getRegionArea(roi_boundary)
tumor_area = spc.msmt.getRegionArea(boundary)
immune_area = spc.msmt.getRegionArea(immune_boundary)
tumor_immune_overlap = boundary.intersection(immune_boundary)
overlap_area = spc.msmt.getRegionArea(tumor_immune_overlap)
print(f"Area of ROI: {roi_area:.2f}")
print(f"Area of main tumor cell region: {tumor_area:.2f}")
print(f"Area of immune cell region: {immune_area}")
print(f"Area of overlap between tumor and immune cell regions: {overlap_area:.2f}")
print(
f"Percentage of tumor region that has overlap with "
f"immune cell region: {overlap_area / tumor_area:.3f}"
)
Area of ROI: 202741300.25
Area of main tumor cell region: 77493721.80
Area of immune cell region: 58152303.47107381
Area of overlap between tumor and immune cell regions: 29934754.20
Percentage of tumor region that has overlap with immune cell region: 0.386
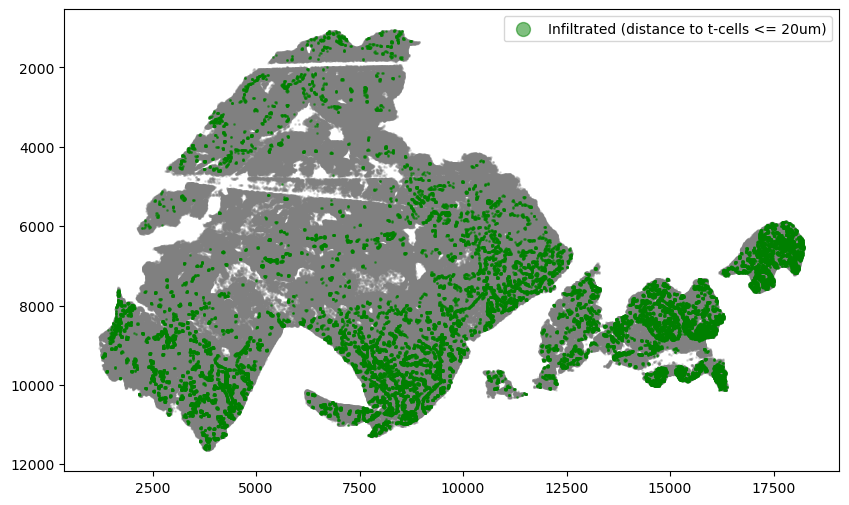
Identify tumor cells that are adjacent to (<=20um) immune cells.
[32]:
dists = spc.msmt.getMinCellTypesDistance(melano, tcells)
adata.obs.loc[
(adata.obs.SOX10_cellRingMask_positive) & (adata.obs.region.isin(["Tumor_ROI", "Tumor"])), "dist"
] = dists
[33]:
threshold = 20
adata.obs["dist_binned"] = adata.obs["dist"] <= threshold
infiltrated = adata.obs[
(adata.obs.SOX10_cellRingMask_positive)
& (adata.obs.region == "Tumor")
& (adata.obs.dist_binned == True)
]
non_infiltrated = adata.obs[
(adata.obs.SOX10_cellRingMask_positive)
& (adata.obs.region == "Tumor")
& (adata.obs.dist_binned == False)
]
fig, ax = plt.subplots(figsize=(10, 6))
ax.invert_yaxis()
region = adata.obs[(adata.obs.region == "Tumor")]
plt.scatter(region["X_centroid"], region["Y_centroid"], s=1, alpha=0.2, color="grey")
plt.scatter(
infiltrated["X_centroid"],
infiltrated["Y_centroid"],
s=1,
alpha=0.5,
color="green",
label=f"Infiltrated (distance to t-cells <= {threshold}um)",
)
plt.legend(markerscale=10)
plt.show()